As the cost of DNA sequencing continues to drop faster than Moore’s Law, there is a growing need for tools that can efficiently analyze larger bodies of sequence data. By mid-2013, sequencing a human genome is expected to cost $1000, at which point this technology enters the realm of routine clinical practice. For example, it is expected that each cancer patient will have their genome and their cancer’s genome sequenced. Assembling and interpreting the short read data produced by sequencers in a timely fashion, however, is a significant challenge, with current pipelines taking thousands of CPU-hours per genome.
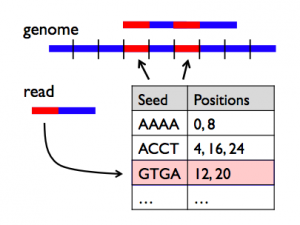
Here, we address the first and most expensive step of this process: aligning reads to a reference genome. We present the Scalable Nucleotide Alignment Program (SNAP), a new aligner that is 10-100x faster and simultaneously more accurate than existing tools like BWA, Bowtie2 and SOAP2. Unlike recent aligners that use graphical processing units (GPUs), SNAP runs on commodity processors. Furthermore, whereas existing fast aligners limit the number and types of differences from the reference genome they allow per read, SNAP supports a rich error model and can cheaply match reads with more differences. This gives it up to 2x lower error rates than current tools and lets it match classes of mutations, such as longer indels, that these tools miss.
SNAP is open source on the SNAP homepage. A technical report on the algorithm is also available.
SNAP is a joint project with Microsoft Research and UC San Francisco.