As data volumes continue to expand, analytics approaches that require exhaustively scanning data sets become untenable. For this reason, we have been developing data organization techniques that make it possible to avoid looking at large volumes of irrelevant data. Our work in this area, which we call “Aggressive Data Skipping” recently got picked up by O’Reilly Radar: Data Today: Behind on Predictive Analytics, Aggressive Data Skipping + More. In this post, I give a brief overview the approach and provide references to more detailed publications.
Data skipping is an increasingly popular technique used in modern analytics databases, including IBM DB2 BLU, Vertica, Spark and many others. The idea is very simple: big data files are partitioned into fairly small blocks of say, 10,000 rows. For each such block we store some metadata, e.g., the min and max of each column. Before scanning each block, a query can first check the metadata and then decide if the block possibly contains records that are relevant to the query. If the metadata indicates that no such records are contained in the block, then the block does not need to be read, i.e, it can be skipped altogether.
In our work we focus on maximizing the amount of data that can be skipped (hence the name “Aggressive Data Skipping”). The key to our approach is Workload Analysis. That is, we observe the queries that are presented to the system over time, and then make partitioning decisions based on what is learned from those observations. Our workload-driven fine-grained partitioning framework re-orders the rows at data loading time.
In order to maximize the chance of data skipping, our research answers the following questions:
- what partitioning method is appropriate for generating fine-grained blocks
- what kind of (concise) metadata can we store for supporting arbitrary filters (e.g., string matching or UDF filters)
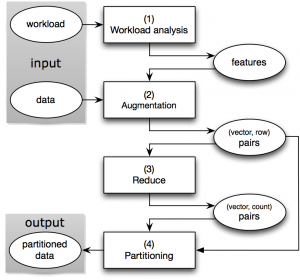
As shown in the figure below, our approach uses the following “W-A-R-P” steps:
- Workload analysis: We extract the frequently recurring filter patterns, which we call the features, from the workload. The workload can be a log of past ad-hoc queries or a collection of query templates from which daily reporting queries are generated.
- Augmentation: For each row, we compute a bit vector based on the features and augment the row with this vector.
- Reduce: We group together the rows with the same bit vectors, since the partitioning decision will be solely based on the bit vectors rather than the actual data rows.
- Partition: We run a clustering algorithm on the bit vectors and generate a partitioning scheme. The rows will be routed to their destination partitions guided by this partitioning scheme.
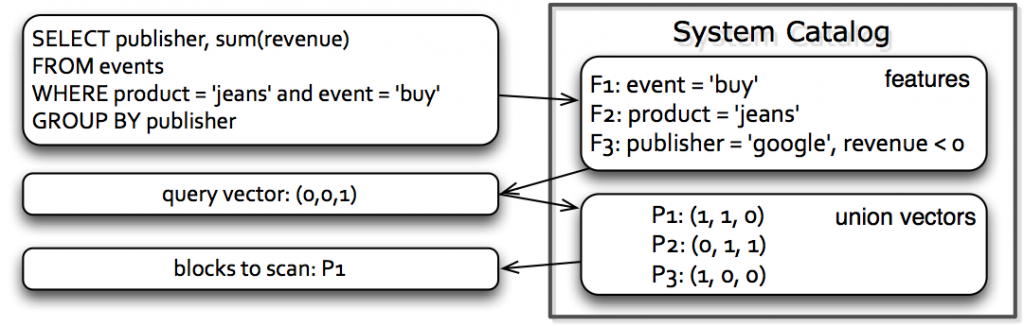
After we have partitioned the data, we store a feature bit vector for each partition as metadata. The following figure illustrates how data skipping works during query execution.
When a query comes, our system first checks which features are applicable for data skipping. With this information, the query processor then goes through the partition-level metadata (i.e., the bit vectors) and decides which partitions can be skipped. This process can work in conjunction with existing data skipping based on min/max.
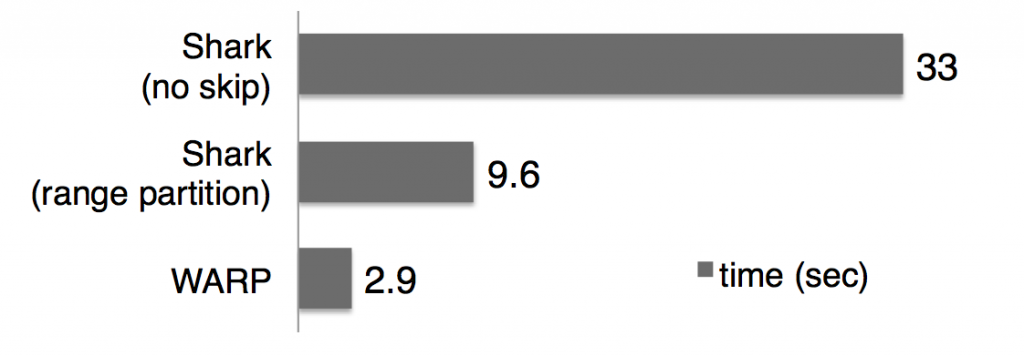
We prototyped this framework on Shark and our experiments with TPC-H and a real-world dataset show speed ups of 2x to 7x. An example result from the TPC-H benchmark (measuring average query response time over 80 TPC-H queries) is shown below.
For more technical details and results, please read our SIGMOD 14 paper, or if you hate formalism and equations, we also gave a demo in VLDB 14. Feel free to send an email to liwen@cs.berkeley.edu for any questions or comments on this project.